The R14 test is a Rapid Whole Genome Sequencing test for acutely unwell children with a likely monogenic disorder, where rapid testing is required to guide immediate clinical management.
We aim to deliver preliminary results via email within 2 weeks of samples arriving in the Exeter Genomics Laboratory.
Please use the links below to jump to the relevant section of this page:
Clinical Application
How to arrange R14 testing for your patient
Consent
Sample Requirements
How is the data analysed?
What if samples from both biological parents aren’t available?
Diagnostic yield
Be aware of what might not be detected
R14 National Educational Multidisciplinary Team (MDT) Meetings
The National Genomic Test Directory for rare and inherited diseases specifies the genomic tests commissioned by the NHS in England for rare and inherited disorders, the technology by which they are available, and the patients who will be eligible to access to a test.
Examples:
Where imminent demise of the patient is predicted then rapid genome sequencing will not be performed.
- Children under 12 months of age with unexplained epilepsy and thought highly likely to have a monogenic cause
- Recent (last 4 months) PICU admission or regular PICU admissions over the last year, epilepsy refractory to treatment and thought highly likely to have a monogenic cause
AND
- Where testing will provide an immediate change to treatment or clinical management for the patient
Exclusion Criteria
- Simple febrile seizures
- Acute or remote symptomatic seizures due to sepsis, haemorrhage, cerebral infarction, hypoxic ischaemic encephalopathy or non-accidental injury
- Structural malformations of the brain where the likely genetic cause is known such as tuberous sclerosis or lissencephaly
The R14 Service is currently commissioned by NHS England to provide testing for 1200 families per year. Inappropriate use of the service diverts resources away from those children who urgently require a molecular diagnosis. We are grateful to all referrers for their help in protecting this valuable resource for these families.
The pathway for R14 testing is outlined below. All R14 requests must be supported by a named Clinical Geneticist and approved by the Exeter R14 Clinical Scientist team prior to discussing the test with the parents and arranging samples. Relevant forms (R14 Record of Discussion (RoD) form and R14 Genome request form) can be found to the right hand side/end of this text in the blue buttons or using the hyperlinked text.
The below workflow can be also be opened on a separate page using this link if you cannot view it clearly.
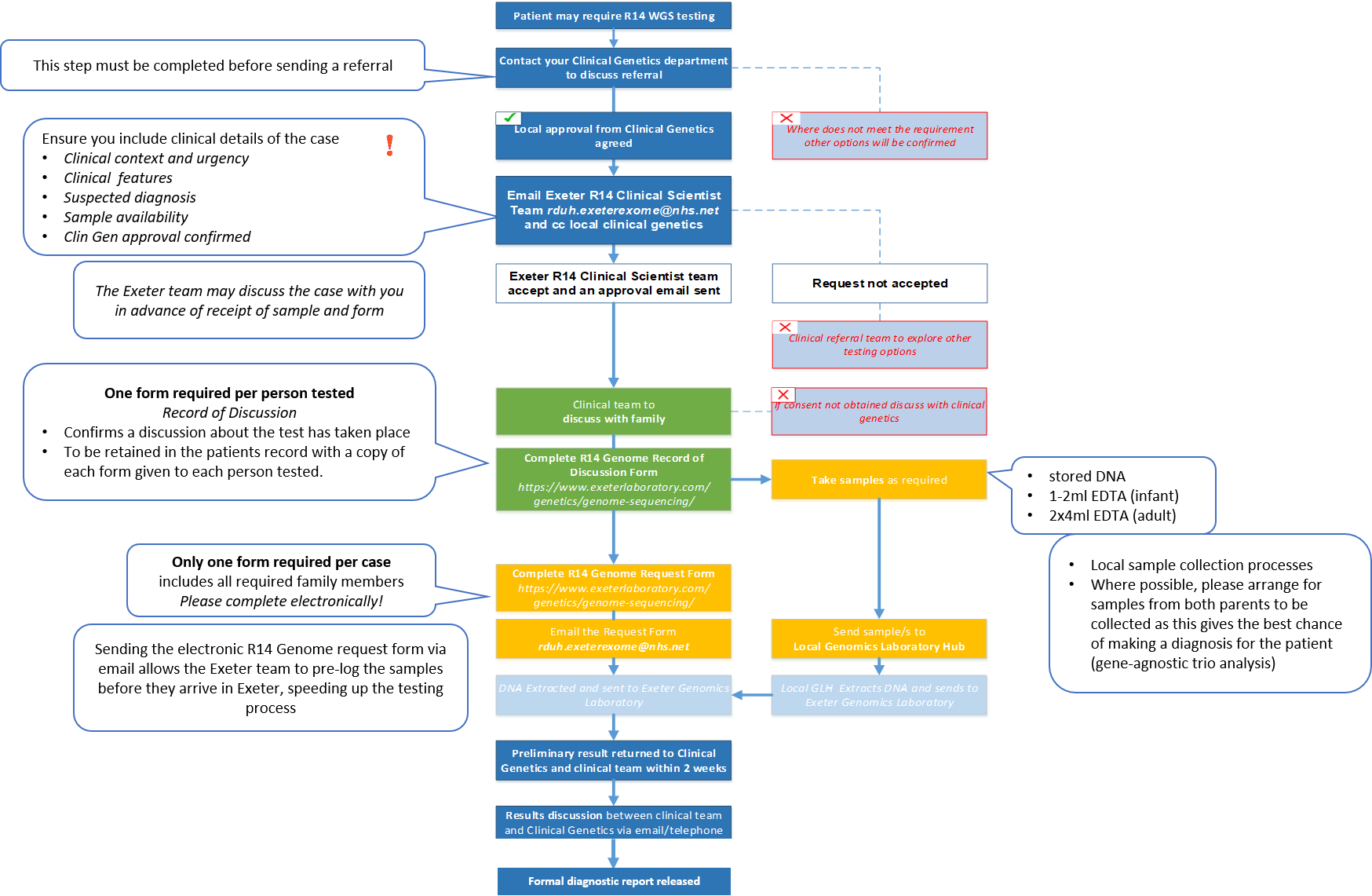
An R14 Record of Discussion form must be completed for each individual being sequenced. This should be stored locally and does not need to be sent to the Exeter Genomics Laboratory. The following points should be discussed by the Clinical team with the parents prior to sequencing:
Peripheral blood samples provide optimal DNA yield and quality for testing over other sample types (saliva, buccal, urine, tissue).
DNA will be extracted from the sample by your local Genomics Laboratory Hub and forwarded to the Exeter Genomics Laboratory for testing. Where possible, at least 1ug DNA should be sent per individual being sequenced. Samples that do not meet these criteria can be discussed with us at rduh.exeterexome@nhs.net.
DNA extracted from amniotic fluid or chorionic villi may also be appropriate for testing where this is the only sample available.
If there is insufficient DNA available from the patient for genome sequencing, it is possible to test for an autosomal recessive aetiology using samples from both parents (Stals et al., 2018 PMID: 29096039).
The genome sequence of each individual is compared to the human reference genome to identify any changes (variants) in the DNA sequence. We all have around 5 million variants in our genomes and it would be impossible for the scientists within the laboratory to review each of these manually. Computational bioinformatics pipelines are used to narrow down this large number of variants to a much smaller list of prioritised variants based on their characteristics, for example how rare they are within the general population.
>90% of R14 tests are performed as a gene-agnostic trio analysis. Here, ‘whole’ genome sequencing is performed for the patient, together with their unaffected parents and an inheritance-based, gene-agnostic strategy is used to look at variants in >20,000 genes. This narrows down the variants using knowledge of how they have been inherited in the child, creating a prioritised variant list for review by the Scientist team:
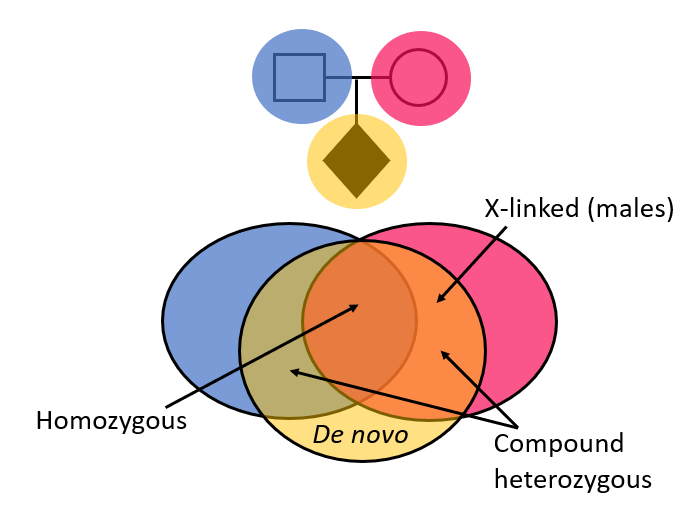
The gene-agnostic trio analysis is not limited to virtual panels of genes known to cause particular clinical presentations.
The prioritised variant list is then reviewed by the analysing scientist using a phenotype-driven approach with the child’s clinical presentation in mind. It is therefore important to include all relevant clinical details/family history on the referral form and share your suspected diagnoses.
The gene-agnostic trio approach does not look for autosomal dominant disorders that have been inherited from a parent. Secondary analysis of the genome sequencing data using virtual gene panels has limited utility after a gene-agnostic trio but may be initiated by the analysing scientist in limited scenarios:
✓ a parent could be mildly/unaffected with the same disorder (reduced penetrance/ parental mosaicism)
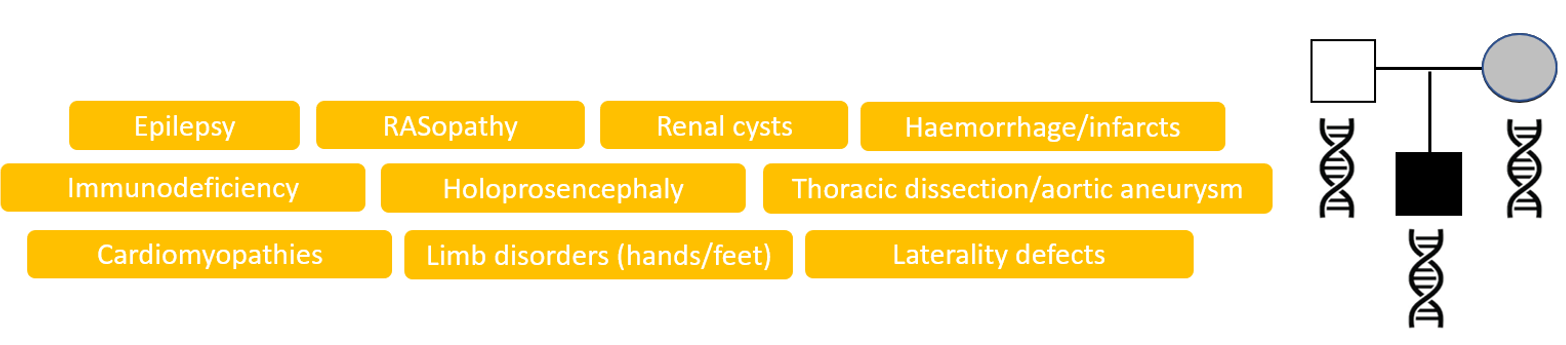
✓ …or there are highly specific clinical features and/or non-genetic investigations that implicate a single gene/small number of genes, which warrants looking for variants that are lower quality/mosaic in the proband or fall outside of our typical region of interest (e.g. deep intronic, previously unreported)

An advantage of using the gene-agnostic trio analysis strategy is that variants in genes that are not yet known to be associated with disease are also included within the prioritised variant list. Where variants are identified in good biological candidate genes, additional cases may be sought via GeneMatcher, which facilitates connections between clinicians and scientists from around the world who share an interest in the same gene or genes. Through this collaborative approach, we have identified several novel disease genes that now have sufficient evidence for their association with disease, warranting their inclusion in virtual gene panels. This enables diagnoses not just for the individual patients tested via R14, but for future patients with variants in these genes that may be tested worldwide (see publications).
Where it is not possible to obtain samples from both biological parents, R14 genome sequencing can be performed as a singleton or duo, however this is a virtual gene panel based analysis. This strategy analyses variants in genes associated with particular clinical indications, for example ‘Early onset or syndromic epilepsy’ or ‘Arthrogryposis’. The virtual gene panels to be applied will be selected from https://panelapp.genomicsengland.co.uk/panels/ and agreed with the referring clinician prior to testing. Where possible, please choose the most specific panel(s) for the patient’s clinical presentation.
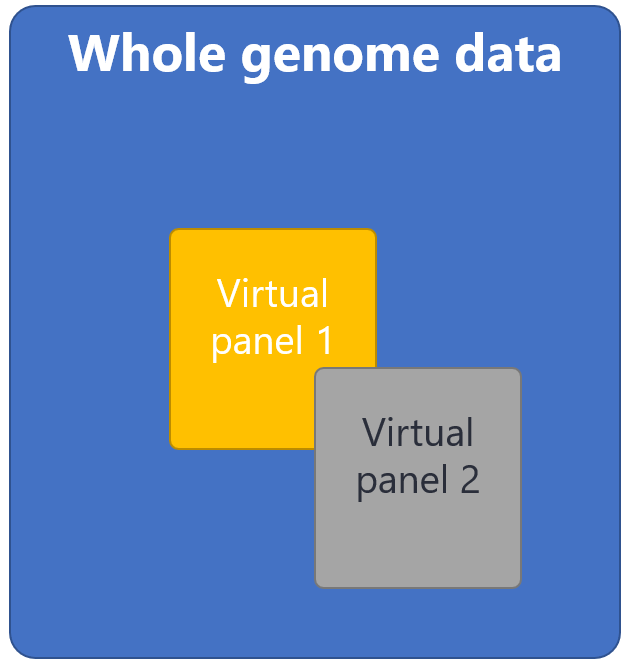
The likelihood of finding a genetic diagnosis by genome sequencing depends upon the patient’s phenotype, i.e. the prior probability that the patient has a monogenic disorder. A successful diagnosis requires that the disease gene is known, there is sufficient coverage (sequencing data) for the gene and that the variant type is detectable by the sequencing technology. The R14 service is aimed at acutely unwell patients with a high likelihood of having a monogenic disorder. Approximately 38-40% of patients having genome sequencing through the R14 service have a diagnosis identified.
R14 genome sequencing is not suitable for the detection of methylation abnormalities and the presence of pseudogenes may prevent the detection of some pathogenic variants. R14 does not currently detect short tandem repeat (STR) disorders. For disorders such as spinal muscular atrophy, congenital myotonic dystrophy, Prader-Willi/Angelman syndromes or congenital central hypoventilation syndrome please request testing from the home Genomic Laboratory Hub using alternative technologies. R14 genome sequencing may detect mitochondrial DNA variants however concurrent mitochondrial DNA testing should be requested from a specialist laboratory if a mitochondrial disorder is suspected. Copy number variants (CNVs) can be detected by R14 genome sequencing, however the presence of a CNV cannot be excluded. Please request urgent microarray (arrayCGH or SNP array) concurrently where appropriate. R14 genome sequencing does not currently detect structural rearrangements e.g. balanced translocations or inversions.
Variants within complex regions of the genome that are not amenable to short read genome sequencing technology may not be detected. To explore how well a particular gene of interest is typically covered by R14 genome sequencing, please use the genome sequencing coverage checker tool.
The R14 genome sequencing analysis pipelines (gene-agnostic trio analysis or virtual panel-based analysis) focus predominantly on exonic regions of the genome*. These are the regions of genes that are retained when introns are removed from the mRNA during splicing. Exonic regions are largely comprised of the regions of DNA that code for proteins, where approximately 85% of pathogenic variants are thought to lie. In addition to the exons, analysis includes the intron-exon boundaries (-50 bp before the exon and +10bp after the exon) as these are well-recognised, important regions where variants may affect splicing.
*Variants that have been previously reported as likely pathogenic or pathogenic in clinical variant databases (ClinVar/HGMD) will be retained by analysis pipelines regardless of their location within the genome (e.g. deep intronic, regulatory regions) using a positive overriding filter.
The Exeter R14 Scientist team hosts a national educational MDT meeting each quarter to share genomics education and service updates. This is enriched with learning from recent R14 cases jointly presented by the Exeter R14 Scientist team and referring clinical teams. Meetings are held via MS Teams and aimed at clinical colleagues within the NHS looking to better understand whole genome sequencing and the R14 service (e.g. Clinical Geneticists, Genomics Counsellors, Neonatologists, Paediatricians, Intensivists, Paediatric Neurologists).
Next meeting:
Wednesday 1st May 2024 13:00-14:30 via MS Teams
Invitations to the meeting will be circulated via email.
If you have a case that you would like to present, please get in touch at rduh.exeterexome@nhs.net.